PakAlumni Worldwide: The Global Social Network
The Global Social Network
Harvard Scientist Debunks Hindu Nationalists "Racial Purity" Myth
Male ancestors of the vast majority of present-day South Asians (Indians, Pakistanis, Bangladeshis) came from West Eurasia, Central Asia and Iran, according to the latest DNA research led by Harvard geneticist Dr. David Reich. Reich's team came to this conclusion after studying the Y-chromosomes of present-day Indians. Some Hindu Indian scientists have used mitochondrial DNA (mtDNA) samples, extracted from the bones of recently discovered ancient skeletal remains of a couple in Rakigarhi in Haryana, to claim the local indigenous origins of all Hindus. Y-chromosomes are passed from father to son while mitochondrial DNA is passed from mother to children. The Harvard team's findings thoroughly debunk Hindu Nationalists' "racial purity" myth similar to that promoted by White Supremacist racists in the West. Reich writes: "The Hindutva ideology that there was no major contribution to Indian culture from migrants from outside South Asia is undermined by the fact that approximately half of the ancestry of Indians today is derived from multiple waves of mass migration from Iran and the Eurasian steppe within the last five thousand years".
 |
| David Reich's "Who We Are" |
Reich's Indian counterparts were highly resistant to the Harvard team findings of foreign origins of modern-day South Asians. Here's an excerpt from David Reich's "Who We Are and How We Got Here":
"Based on their own mitochondrial DNA studies, it was clear to them (Indians) that the great majority of mitochondrial DNA lineages present in India today had resided in the subcontinent for many tens of thousands of years.They did not want to be part of a study that suggested a major West Eurasian incursion into India without being absolutely certain as to how the whole-genome data could be reconciled with their mitochondrial DNA findings. They also implied that the suggestion of a migration from West Eurasia would be politically explosive. They did not explicitly say this, but it had obvious overtones of the idea that migration from outside India had a transformative effect on the (South Asian) subcontinent".
"To keep up the purity of the Race and its culture, Germany shocked the world by her purging the country of the Semitic races -- the Jews. Race pride at its highest has been manifested here. Germany has also shown how well-nigh impossible it is for races and cultures, having differences going to the root, to be assimilated into one united whole, a good lesson for us in Hindusthan to learn and profit by."
"Groups of traditionally higher social status in the Indian caste system typically have a higher proportion of ANI ancestry than those of traditionally lower social status, even within the same state of India where everyone speaks the same language. For example, Brahmins, the priestly caste, tend to have more ANI ancestry than the groups they live among, even those speaking the same language. Although there are groups in India that are exceptions to these patterns, including well-documented cases where whole groups have shifted social status, the findings are statistically clear, and suggest that the ANI-ASI mixture in ancient India occurred in the context of social stratification".
 |
| South Asian Ancestry. Source: Arain Gang |
 |
| 1901 Indian Census of UP Muslims |
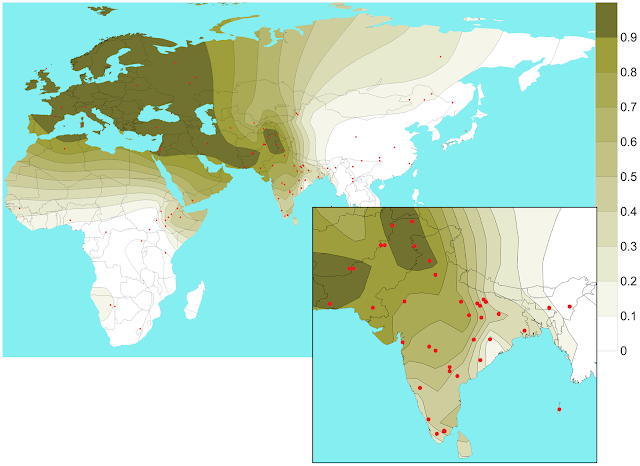 |
| Light Skin Gene Distribution. Source: PLOS Genetics |
 |
| 1901 India Census UP Hindu Population By Castes |
World Values Survey Finds Indians Most Racist
Indians Admire Israel and Hitler
Caste Apartheid in India
Religion, Caste and Politics in India by Christophe Jaffrelot
Mahatma Gandhi and His Struggle With India
Who Killed Karkare?
Procrastinating on Hindutva Terror
India's Guantanamos and Abu Ghraibs
Hindutva Government in Israeli Exile?
Growing US-India Military Ties Worry Pakistan
The 21st Century Challenges For Resurgent India
-
Comment by Riaz Haq on February 3, 2021 at 6:37pm
-
The Light Skin Allele of SLC24A5 in South Asians and Europeans Shares Identity by Descent
https://journals.plos.org/plosgenetics/article?id=10.1371/journal.p...
Skin pigmentation is one of the most variable phenotypic traits in humans. A non-synonymous substitution (rs1426654) in the third exon of SLC24A5 accounts for lighter skin in Europeans but not in East Asians. A previous genome-wide association study carried out in a heterogeneous sample of UK immigrants of South Asian descent suggested that this gene also contributes significantly to skin pigmentation variation among South Asians. In the present study, we have quantitatively assessed skin pigmentation for a largely homogeneous cohort of 1228 individuals from the Southern region of the Indian subcontinent. Our data confirm significant association of rs1426654 SNP with skin pigmentation, explaining about 27% of total phenotypic variation in the cohort studied. Our extensive survey of the polymorphism in 1573 individuals from 54 ethnic populations across the Indian subcontinent reveals wide presence of the derived-A allele, although the frequencies vary substantially among populations. We also show that the geospatial pattern of this allele is complex, but most importantly, reflects strong influence of language, geography and demographic history of the populations. Sequencing 11.74 kb of SLC24A5 in 95 individuals worldwide reveals that the rs1426654-A alleles in South Asian and West Eurasian populations are monophyletic and occur on the background of a common haplotype that is characterized by low genetic diversity. We date the coalescence of the light skin associated allele at 22–28 KYA. Both our sequence and genome-wide genotype data confirm that this gene has been a target for positive selection among Europeans. However, the latter also shows additional evidence of selection in populations of the Middle East, Central Asia, Pakistan and North India but not in South India.
-------
We also studied the geospatial pattern of rs1426654-A allele frequencies at the global level using 2763 subjects from previously published data (Table S7) and 1446 individuals from the present study (Table S5). The isofrequency map illustrates high frequencies of the rs1426654-A allele in Europe, Middle East, Pakistan, moderate to high frequencies in Northwest and Central Asia, while being almost absent in East Asians and Africans with notable exceptions in Bantu (Southwest), San, Mandeka, and Ethiopians (Table S7, Figure 2).
-----------
Hence, these observations confirm that SLC24A5 has been under strong selective pressure in Europeans. In addition to this, we also performed haplotype-based selection tests based on genome-wide data (see Materials and Methods) of 1035 individuals including 145 Indians. XP-EHH scores demonstrated that SLC24A5 ranks among the top 10 candidate genes for positive selection in Europe, Middle East and Pakistan, and among the top 1% in Central Asia, Iran and North India (Table S13). Likewise, scores from our iHS analysis had significant empirical p-values for Central Asia and North India (Table S13). It is interesting to note that both of our haplotype-based selection tests demonstrated evidence of positive selection in North Indians, but no such evidence of positive selection was found in South Indians (Table S13). The difference in detecting selection signals from genotype and sequence data has also been pinpointed in a previous study [31].
-
Comment by Riaz Haq on February 4, 2021 at 9:19am
-
Two new genetic studies upheld Indo-Aryan migration. So why did Indian media report the opposite?
Science is proving India's incredible diversity. But this clashes directly with Hindutva’s racially nativist understanding of the subcontinent.
https://scroll.in/article/936872/two-new-genetic-studies-upheld-ary...
The Economic Times reported that the research raises doubts over the “long-held theory of Aryan invasion or migration into South Asia”. Amar Ujala, one of India’s largest Hindi newspapers, was more emphatic: “The Aryan invasion theory proved completely false; India is the guru of South Asia.”
The theory of the Aryan invasion (or migration) was first put forward by Western scholars during the colonial age. It maintained that a race of European or Central Asian “Aryans” swept into the subcontinent displacing the indigenous Indus Valley Civilisation. These Aryans were said to have introduced key elements of Indian culture such as the Sanskrit language – which gave rise to the Indo-Aryan branch of languages spoken all across north, west and east India today – as well as the Vedas, the foundational texts of Hinduism.
This went against Hindutva’s own imagination of India, in which all significant cultural development was held to be indigenous.
Some of what the term “Aryan” once referred to has been proved to be scientifically inaccurate. The Nazis, for example, mistook what is a language grouping to be a racial one. However, much of the Indian media did not bother to explain that the new research actually upheld the theory that people with European Steppe ancestry had brought the Indo-Aryan language branch to India – not overturned it.
In contrast, the media in the West (with no political dog in this fight) communicated this fact rather well. People of Steppe-pastoralist ancestry likely “brought horses and the Indo-European languages now spoken on the subcontinent,” reported the Atlantic. The Smithsonian.com website of the American museum group wrote, “Indo-European languages may have reached South Asia via Central Asia and Eastern Europe during the first half of the 1000s BC.”
These results were not only misinterpreted in the media, they also led to a split in how the authors of the landmark studies and other genetic scientists interpreted them. Vasant Shinde, co-author on both studies, put out a press release on September 6 where he argued that the new data “completely sets aside the Aryan Migration/Invasion Theory” and also proves that the “Harappans were the Vedic people”.
When Scroll.in spoke to Shinde, he explained his point further. “This is not a migration but a movement of people,” Shinde argued. “And the movement from the Steppe is not large.”
Shinde also disagreed with the linguistic conclusions in the research, claiming that they were not based on any scientific proof. “The Harappans were speaking Sanskrit since they were so advanced,” Shinde told Scroll.in.
American geneticist and science writer Razib Khan did not agree with Shinde’s conclusions. “This research points strongly to the fact that Aryans migrated to the Indian subcontinent,” said Khan. “Steppe ancestry is found in almost every group in India. And Steppe ancestry maps to the spread of Indo-Aryan language migration”.
What about the Shinde’s conclusion that the people of the Indus Valley Civilisation were the same as the Vedic people? “I at least cannot make such an interpretation,” Vagheesh Narasimhan, co-author of the Science study told Scroll.in. “This proposition makes jumps that I am not comfortable with.”
Another co-author on the Science paper, Niraj Rai chose his words carefully when it came to Shinde’s claim of equating the Indus Valley Civilisation with the culture that authored the Vedas. “This is not my statement; I don’t agree with this statement,” said the geneticist.
-
Comment by Riaz Haq on February 4, 2021 at 9:20am
-
Two new genetic studies upheld Indo-Aryan migration. So why did Indian media report the opposite?
Science is proving India's incredible diversity. But this clashes directly with Hindutva’s racially nativist understanding of the subcontinent.
https://scroll.in/article/936872/two-new-genetic-studies-upheld-ary...
Another co-author on the Science paper, Niraj Rai chose his words carefully when it came to Shinde’s claim of equating the Indus Valley Civilisation with the culture that authored the Vedas. “This is not my statement; I don’t agree with this statement,” said the geneticist.
Nick Patterson, another co-author, and one of the main movers along with geneticist David Reich of the endeavour to genetically decode South Asian origins had much the same point to make while speaking to Scroll.in: “While I am always willing to listen, I disagree with Dr Shinde that the people of the Indus Valley spoke an Indo-European language.”
The study of the genetics of ancient humans, using DNA to do the work of archeologists and historians in explaining our past, was pioneered at Harvard University by geneticist David Reich. However, till now, India has been one of the dark spots in this field of study. This is because, as Nick Patterson, a co-author on the two new papers and a close associate of Reich, told Scroll.in, “Genetic material survives best in cool and dry climates. And two words I would not use to describe India are cool and dry.”
However, researchers managed to extricate enough DNA from the ear bone of a woman who lived in the Indus Valley Civilisation 4,500 years ago from an archeological site in Rakhigarhi, Haryana. This DNA was analysed by Reich’s team in one paper published in Cell. Another analysis by Reich’s team in Science took a macro view, analysing genetic data from 523 ancient people spanning 8,000 years across Central and South Asia right up to the European Steppe – the largest study of ancient human DNA.
The picture that emerges is one of diverse origin for the modern South Asian population. The main building blocks at the time of the Bronze Age (around four millennia ago) are the Ancient Ancestral South Indians, the people of the Indus Valley Civilisation and a significant migration from the Pontic Steppe. None of these people exist today but it is their mixing that caused most of the modern Indian population to be formed.
Of these, the Ancient Ancestral South Indians are probably the least studied and were present across parts of the subcontinent that did not fall under the Indus Valley Civilisation. Their closest modern-day relatives are the tribes of the Andaman Islands.
Thanks to the Cell paper released on September 5, we now know that the people of the Indus Valley had no Steppe DNA. They mainly had a mixture of Iranian-farmer-related DNA as well as some DNA from Ancient Ancestral South Indians.
The Steppe population came in from grasslands in Eastern Europe corresponding to modern-day Ukraine, Russia and Kazakhstan. The genetic research identifies that this Steppe ancestry burst into India during a “narrow time window” dated between 2,000 BC and 1,500 BC.
Once these Steppe people entered India, a great churning ensued. They mixed with the Indus Valley people to create what is now called the Ancestral North Indian grouping. However, a significant portion of the people of the Indus Valley Civilisation were pushed south when the Steppe people entered. They then mixed with the Ancient Ancestral South Indians to form a group known as the Ancestral South Indian population.
For the next 2,000 years, Indians mixed freely. As a result, most modern South Asians are some mix of Ancestral North Indian and Ancestral South Indian. However, this great churning stopped around 1,900 years ago when Indian society calcified into thousands of endogamous groups who do not intermarry across caste lines – a societal structure maintained till today.
-
Comment by Riaz Haq on February 4, 2021 at 9:20am
-
Two new genetic studies upheld Indo-Aryan migration. So why did Indian media report the opposite?
Science is proving India's incredible diversity. But this clashes directly with Hindutva’s racially nativist understanding of the subcontinent.
https://scroll.in/article/936872/two-new-genetic-studies-upheld-ary...
There are however some exceptions to this narrative. The Bengalis and Mundas, an Adivasi people of eastern India, “have significant amounts of ancestry from South East Asia”, noted Razib Khan, and cannot be explained using this Ancestral North Indian-Ancestral South Indian model.
How this explains modern India
Perhaps the biggest learning from this genetic research is that it explains the various languages South Asians speak. “It is clear that the movement of people mirrors the information we have from linguistics on how different features of language families are shared between them,” explained Vagheesh Narasimhan of the Department of Genetics at Harvard Medical School, who has contributed to the research.
The main theory to which Narasimhan is referring deals with how a single language family, the Indo-European family stretches all the way from Britain to Bangladesh and encompasses more than half of the modern world’s population. It counts amongst its members ancient heavyweights such as Sanskrit, Greek, Roman and Pali as well as modern tongues like English, Persian and Hindi.
The people who spread the Indo-European language family across Eurasia are the same Steppe pastoralists who are key constituents in making up the modern Indian population. As the Science paper states, its results provide “evidence for the theory that these [Indo-European] languages spread from the Steppe”.
In South Asia, the Indo-European language family bought in by the Steppe people forked to give rise to the Indo-Aryan daughter branch. The first Indo-Aryan language in South Asia was Vedic Sanskrit, the language of the Rig Veda. In present-day South Asia, around 1.3 billion people speak an Indo-Aryan language. Each of the modern states of India, Pakistan, Bangladesh and Sri Lanka use an Indo-Aryan language as their official language.
Much of this Steppe ancestry is male, the research shows. This means that Steppe migrants “were more successful at competing for local mates than men from the local groups” – which tells us something about the aggressive nature of Indo-Aryan migration into India. The Science paper concludes that there was an “asymmetric social interaction between descendants of Steppe pastoralists and peoples of the Indus Periphery Cline [Indus Valley Civilisation]”.
In simpler language, David Reich explains that the preponderence of male Steppe DNA means that this encounter between the Steppe pastoralists and the people of the Indus Valley Civilisation “cannot have been entirely friendly”.
This male bias is standard for Indo-European migration. In fact, when these Steppe pastoralists reached Europe, Reich’s research found an even larger proportion of male Steppe genes. In large parts of Western Europe, Steppe migrants almost completely displaced local males in a short time span, leading to one Danish archeologist postulating that the coming of these Indo-European speakers “must have been a kind of genocide”.
This pattern, wrote David Reich in his 2018 book Who We Are and How We Got Here, “is exactly what one would expect from an Indo-European-speaking people taking the reins of political and social power 4,000 years ago and mixing with the local peoples in a stratified society, with males from the groups in power having more success in finding mates than those from the disenfranchised groups”.
-
Comment by Riaz Haq on February 4, 2021 at 10:03am
-
2,000-Year-Old #Buddhist Site Unearthed in #Swat #Pakistan. The site housed an “educational institution” that included a schooling area, an assembly hall, meditation shrines called stupas, and viharas, or small living quarters for monks. https://news.yahoo.com/2-000-old-buddhist-unearthed-012748169.html?... via @YahooNews
Archaeologists uncovered a 1,900 to 2,000-year-old Buddhist site on cattle grazing land in Pakistan’s Swat District, Radio Free Europe/Radio Liberty (RFE/RL) reports.
According to lead archaeologist Saqib Raza, the site housed an “educational institution” that included a schooling area, an assembly hall, meditation shrines called stupas, and viharas, or small living quarters for monks. Raza said the complex is from the Kushan era and its structures were damaged over time by various groups that came to settle in the area, eventually including the Hindu Shahi dynasty and Muslims.
According to RFE/RL’s reporter on the ground, work on the site started in December of 2020 and was expected to continue until March of 2021, in preparation for the site to eventually open to visitors. Credit: Radio Free Europe/Radio Liberty via Storyful
-
Comment by Riaz Haq on February 4, 2021 at 6:46pm
-
YORKSHIRE, ENGLAND—Could men from Central Asia have brought Indo-European languages to India thousands of years ago? Live Science reports that archaeogeneticist Martin Richards of the University of Huddersfield and his colleagues conducted an analysis of genetic material collected from modern populations living in India, where ancient DNA rarely survives in the hot climate. The data from the study suggest that multiple waves of people migrated into the subcontinent from Anatolia, the Caucasus, and Iran over the past 20,000 years. In particular, Richards and his colleagues say between 4,000 and 3,800 years ago, the Y-chromosome subtype associated with men of the Yamnaya culture appeared in Indian populations. The Yamnaya lived between the Caspian Sea and the Black Sea, are known for their pit graves and wheeled horse chariots, and spoke a precursor of Indo-European languages. The Yamnaya subgroup is now carried by about 17.5 percent of Indian men—more often in the north than in the south. So, did Yamnaya warriors conquer northern India, or perhaps migrate there and have large families? “It’s very easy for Y-chromosome composition to change very quickly,” Richards said. “Just because individual men can have a lot more children than women can.” To read more about Bronze Age Indo-European cultures, go to "The Wolf Rites of Winter."
https://www.archaeology.org/news/5715-170706-india-genetic-study
-----------------
One of the major factors that has still kept the origin of the Indian caste system obscure is the unresolved question of the origin of Y-haplogroup R1a1*, at times associated with a male-mediated major genetic influx from Central Asia or Eurasia, which has contributed to the higher castes in India. Y-haplogroup R1a1* has a widespread distribution and high frequency across Eurasia, Central Asia and the Indian subcontinent, with scanty reports of its ancestral (R*, R1* and R1a*) and derived lineages (R1a1a, R1a1b and R1a1c). To resolve these issues, we screened 621 Y-chromosomes (of Brahmins occupying the upper-most caste position and schedule castes/tribals occupying the lower-most positions) with 55 Y-chromosomal binary markers and seven Y-microsatellite markers and compiled an extensive dataset of 2809 Y-chromosomes (681 Brahmins, and 2128 tribals and schedule castes) for conclusions. A peculiar observation of the highest frequency (up to 72.22%) of Y-haplogroup R1a1* in Brahmins hinted at its presence as a founder lineage for this caste group. Further, observation of R1a1* in different tribal population groups, existence of Y-haplogroup R1a* in ancestors and extended phylogenetic analyses of the pooled dataset of 530 Indians, 224 Pakistanis and 276 Central Asians and Eurasians bearing the R1a1* haplogroup supported the autochthonous origin of R1a1 lineage in India and a tribal link to Indian Brahmins. However, it is important to discover novel Y-chromosomal binary marker(s) for a higher resolution of R1a1* and confirm the present conclusions.
https://www.nature.com/articles/jhg20082
-
Comment by Riaz Haq on February 4, 2021 at 6:58pm
-
Haplogroup R1a (Y-DNA) in Pakistan
https://www.eupedia.com/europe/Haplogroup_R1a_Y-DNA.shtml
Haplogroup R* originated in North Asia just before the Last Glacial Maximum (26,500-19,000 years before present). This haplogroup has been identified in the 24,000 year-old remains of the so-called "Mal'ta boy" from the Altai region, in south-central Siberia (Raghavan et al. 2013). This individual belonged to a tribe of mammoth hunters that may have roamed across Siberia and parts of Europe during the Paleolithic. Autosomally this Paleolithic population appears to have passed on its genes mostly to the modern populations of Europea and South Asia, the two regions where haplogroup R also happens to be the most common nowadays (R1b in Western Europe, R1a in Eastern Europe, Central and South Asia, and R2 in South Asia).
The series of mutations that made haplogroup R1* evolve into R1a probably took place during or soon after the Last Glacial Maxium. Little is know for certain about R1a's place of origin. Some think it might have originated in the Balkans or around Pakistan and Northwest India, due to the greater genetic diversity found in these regions. The diversity can be explained by other factors though. The Balkans have been subject to 5000 years of migrations from the Eurasian Steppes, each bringing new varieties of R1a. South Asia has had a much bigger population than any other parts of the world (occasionally equalled by China) for at least 10,000 years, and larger population bring about more genetic diversity. The most likely place of origin of R1a is Central Asia or southern Russia/Siberia.
-------
The Indo-Iranian migrations progressed further south across the Hindu Kush. By 1700 BCE, horse-riding pastoralists had penetrated into Balochistan (south-west Pakistan). The Indus valley succumbed circa 1500 BCE, and the northern and central parts of the Indian subcontinent were taken over by 500 BCE. Westward migrations led Old Indic Sanskrit speakers riding war chariots to Assyria, where they became the Mitanni rulers from circa 1500 BCE. The Medes, Parthians and Persians, all Iranian speakers from the Andronovo culture, moved into the Iranian plateau from 800 BCE. Those that stayed in Central Asia are remembered by history as the Scythians, while the Yamna descendants who remained in the Pontic-Caspian steppe became known as the Sarmatians to the ancient Greeks and Romans.
The Indo-Iranian migrations have resulted in high R1a frequencies in southern Central Asia, Iran and the Indian subcontinent. The highest frequency of R1a (about 65%) is reached in a cluster around Kyrgyzstan, Tajikistan and northern Afghanistan. In India and Pakistan, R1a ranges from 15 to 50% of the population, depending on the region, ethnic group and caste. R1a is generally stronger is the North-West of the subcontinent, and weakest in the Dravidian-speaking South (Tamil Nadu, Kerala, Karnataka, Andhra Pradesh) and from Bengal eastward. Over 70% of the Brahmins (highest caste in Hindusim) belong to R1a1, due to a founder effect.
Maternal lineages in South Asia are, however, overwhelmingly pre-Indo-European. For instance, India has over 75% of "native" mtDNA M and R lineages and 10% of East Asian lineages. In the residual 15% of haplogroups, approximately half are of Middle Eastern origin. Only about 7 or 8% could be of "Russian" (Pontic-Caspian steppe) origin, mostly in the form of haplogroup U2 and W (although the origin of U2 is still debated). European mtDNA lineages are much more common in Central Asia though, and even in Afghanistan and northern Pakistan. This suggests that the Indo-European invasion of India was conducted mostly by men through war. The first major settlement of Indo-Aryan women was in northern Pakistan, western India (Punjab to Gujarat) and northern India (Uttar Pradesh), where haplogroups U2 and W are the most common today.
-
Comment by Riaz Haq on February 4, 2021 at 7:17pm
-
Where West Meets East: The Complex mtDNA Landscape of the Southwest and Central Asian Corridor
https://www.sciencedirect.com/science/article/pii/S0002929707643523
The southwestern Asian corridor is a wide geographical area that extends from Anatolia and the trans-Caucasus area through the Iranian plateau to the Indo-Gangetic plains of Pakistan and northwestern India. This region is characterized by a patchwork of different physical-anthropology types with complex boundaries and gradients and by the coexistence of several language families (e.g., Indo-European, Turkic, and Sino-Tibetan) as well as relict linguistic outliers. The southwestern Asian corridor, located at the crossroads of major population expansions, was the first portion of Eurasia to be inhabited by the Homo sapiens sapiens population(s) that left Africa ∼60,000 years before the present (YBP) (Tishkoff et al. 1996; Watson et al. 1997; Quintana-Murci et al. 1999), and from this region modern humans migrated to the rest of the world. Although Paleolithic and Mesolithic people left their mark in the area, major prehistorical and historical events with possible genetic consequences occurred during the Neolithic period and later. Important agricultural developments occurred in the eastern horn of the Fertile Crescent ∼8,000 YBP, notably in Elam (southwestern Iran). The highly urban Elamite civilization had close contacts with Mesopotamians but exhibited an extensive differentiation from the rest of the Fertile Crescent populations, including a language that is thought to belong to the Dravidian family. It is hypothesized that the proto-Elamo-Dravidian language (McAlpin 1974, 1981), spoken by the Elamites in southwestern Iran, spread eastwards with the movement of farmers from this region to the Indus Valley and the Indian subcontinent (Cavalli-Sforza et al. 1994; Cavalli-Sforza 1996; Renfrew 1996). Starting ∼5,000 YBP, animal domestication, particularly the horse, gave the inhabitants of the Central Asian steppes the opportunity to expand geographically in different directions (Zvelebil 1980). These Central Asian nomads, probably from the Andronovo and Srubnaya cultures, migrated through Iran and Afghanistan, reaching Pakistan and India, and their arrival is contemporaneous with the decline of the strong agricultural South Asian civilizations, such as the Harappans.
------------
The approximate location of the 23 populations from which the 910 mtDNAs were sampled is shown in figure 1. Each sample comprises unrelated healthy donors from whom appropriate informed consent was obtained. For the preliminary part of the study, 208 individuals from three different geographic regions were analyzed: 58 individuals from the Caucasus, 50 from Turkey and 100 from southeastern Pakistan. The three samples were heterogeneous; the sample from the Caucasus region was made up of three different ethnic groups: Georgians, Balkarians, and Chechens. The sample from Turkey was collected mainly in Konya (Anatolia). The Pakistani sample was collected in Karachi and comprised mainly Sindhis, who are a mix of tribes of different religions and ethnicities from the southeastern province of Sindh. The extended population sample included 702 individuals from 20 different populations living in the Iranian plateau, the Indus Valley, the Karakorum and Hindu Kush mountains, and Central Asia. Further details of the whole sample collection are reported in table 1 and in the work of Wells et al. (2001) and Qamar et al. (2002). The term “Makrani” refers to the so-called “Negroid Makrani” population living in the Makran coast of Baluchistan, distinct from the Makrani Baluch population, which is not considered in this study.
-
Comment by Riaz Haq on February 4, 2021 at 7:47pm
-
Where West Meets East: The Complex mtDNA Landscape of the Southwest and Central Asian Corridor
https://www.sciencedirect.com/science/article/pii/S0002929707643523
Traces of Recent and Sexually Asymmetrical Events
The phylogeographical cross-comparison of mtDNA and Y-chromosomal data is very useful for tracing differential male and female histories. Some populations studied here (Iranian, Pakistani, and Central Asian) have been analyzed previously for Y-chromosomal variation (Quintana-Murci et al. 2001; Qamar et al. 2002; Zerjal et al. 2002). In most cases, mtDNA variation is in good agreement with the Y-chromosomal data, suggesting that the patterns reflect general population processes. A good, although surprising, example of concordance between the two systems is the Hazara, who claim to be the direct male-line descendants of Genghis Khan’s army. The presence and time depth of the Y-chromosomal haplogroup C* (xC3c) in the Hazara, along with its absence from neighboring populations, has been interpreted as the genetic legacy of Genghis Khan and his male relatives (Qamar et al. 2002; Zerjal et al. 2003). Our results indicate that the Hazara are also characterized by very high frequencies of eastern Eurasian mtDNAs (35%, table 2, fig. 1), which are virtually absent from bordering populations, suggesting that the male descendants of Genghis Khan, or other Mongols, were accompanied by women of East Asian ancestry.
In contrast to the parallelism between mtDNA and Y-chromosomal data in most populations, the Parsis and the Makrani both show a sharp contrast between these loci. The Parsis live in southeastern Pakistan, and historical records indicate an Iranian origin (Nanavutty 1997). These followers of the prophet Zoroaster started their migration from Iran in the 7th century a.d., settling in the northwestern Indian province of Gujarat around 900 a.d. and eventually moving to Mumbai in India and Karachi in Pakistan. Y-chromosome data show that they resemble Iranian populations rather than their neighbors in Pakistan: an admixture estimate of 100% from Iran was obtained (Qamar et al. 2002), supporting the historical records. However, when the Parsi mtDNA pool was compared with those of the Iranians and Gujaratis (their putative parental populations), a strong contrast with the Y-chromosomal data emerged. About 60% of their maternal gene pool belongs to South Asian haplogroups, which make up only 7% of the combined Iranian sample (table 2). The very high frequency of haplogroup M among the Parsis (55%), similar to those of Indian populations and much higher than that of the combined Iranian sample (1.7%), highlights their close affinities with India (fig. 6). Our results lead to an admixture estimate of 100% from Gujarat and provide a strong contrast between the maternal and paternal components of this population. Although the small population size of the Parsis (a few thousand) may have distorted haplogroup frequencies in this population, diversity of both Y-chromosome and mtDNA lineages remains high, making a strong drift effect unlikely. Our results therefore support a male-mediated migration of the ancestors of the present-day Parsi population from Iran to India, where they admixed with local females, or directional mating in Gujarat between Iranian males and local women, leading ultimately to the loss of mtDNAs of Iranian origin.
-
Comment by Riaz Haq on February 5, 2021 at 8:47am
-
The fusion of these highly different populations into today’s West Eurasians is vividly evident in what might be considered the classic northern European look: blue eyes, light skin, and blond hair. Analysis of ancient DNA data shows that western European hunter-gatherers around eight thousand years ago had blue eyes but dark skin and dark hair, a combination that is rare today.33 The first farmers of Europe mostly had light skin but dark hair and brown eyes—thus light skin in Europe largely owes its origins to migrating farmers.34 The earliest known example of the classic European blond hair mutation is in an Ancient North Eurasian from the Lake Baikal region of eastern Siberia from seventeen thousand years ago.35 The hundreds of millions of copies of this mutation in central and western Europe today likely derive from a massive migration into the region of people bearing Ancient North Eurasian ancestry, an event that is related in the next chapter.36Since the original excavations at Harappa, the “Aryan invasion theory” has been seized on by nationalists in both Europe and India, which makes the idea difficult to discuss in an objective way. European racists, including the Nazis, were drawn to the idea of an invasion of India in which the dark-skinned inhabitants were subdued by light-skinned warriors related to northern Europeans, who imposed on them a hierarchical caste system that forbade intermarriage across groups. To the Nazis and others, the distribution of the Indo-European language family, linking Europe to India and having little impact on the Near East with its Jews, spoke of an ancient conquest moving out of an ancestral homeland, displacing and subjugating the peoples of the conquered territories, an event that they wished to emulate.9 Some placed the ancestral homeland of the Indo-Aryans in northeast Europe, including Germany. They also adopted features of Vedic mythology as their own, calling themselves Aryans after the term in the Rig Veda, and appropriating the swastika, a traditional Hindu symbol of good fortune.10 The Nazis’ interest in migrations and the spread of Indo-European languages has made it difficult for serious scholars in Europe to discuss the possibility of migrations spreading Indo-European languages.11 In India, the possibility that the Indus Valley Civilization fell at the hands of migrating Indo-European speakers coming from the north is also fraught, as it suggests that important elements of South Asian culture might have been influenced from the outside.
Reich, David. Who We Are and How We Got Here (p. 150). Knopf Doubleday Publishing Group. Kindle Edition.
Reich, David. Who We Are and How We Got Here (p. 121). Knopf Doubleday Publishing Group. Kindle Edition.
Twitter Feed
Live Traffic Feed
Sponsored Links
South Asia Investor Review
Investor Information Blog
Haq's Musings
Riaz Haq's Current Affairs Blog
Please Bookmark This Page!
Blog Posts
Trump Leads America into an Unpopular War in the Middle East!
President Donald Trump joined Israel in yet another war of choice in the Middle East last week. Polls conducted in the United States immediately after the start of the Iran war show that the majority of Americans do not support it. A YouGov snap poll fielded Saturday — the day of the strikes — found 34% of Americans approve of the U.S. attacks on Iran, with 44%…
ContinuePosted by Riaz Haq on March 3, 2026 at 10:00am — 3 Comments
India-Israel Axis Threatens Peace in South Asia
The bonhomie between Israeli Prime Minister Netanyahu, an indicted war criminal, and Indian Prime Minister Narendra Modi, accused of killing thousands of Muslims, was on full display this week in Israel. Both leaders committed to supporting the Afghan Taliban regime which is accused of facilitating cross-border terrorist attacks by the TTP in Pakistan. Mr. Modi was warmly welcomed by…
ContinuePosted by Riaz Haq on February 27, 2026 at 10:45am — 2 Comments
© 2026 Created by Riaz Haq.
Powered by
![]()
You need to be a member of PakAlumni Worldwide: The Global Social Network to add comments!
Join PakAlumni Worldwide: The Global Social Network